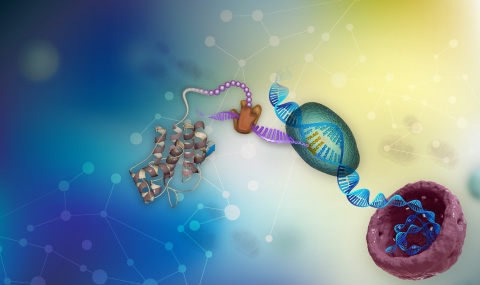
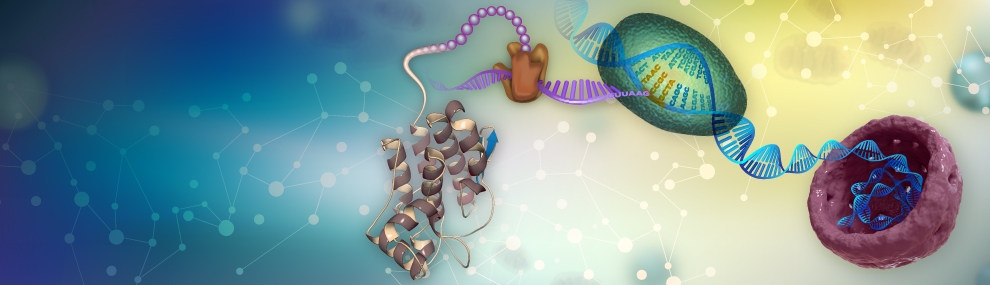
We take a genome wide and a systems-level approach to ask basic biology questions on the protein translation process, particularly its efficiency and fidelity (Man & Pilpel, Nature genetics 2007). We combine computational biology, modeling and experiments to understand translation at the proteome level. We discovered the “Ramp” in protein translation – a universal mechanism that appears to allow efficient translation especially in highly expressed proteins (Tuller et al. Cell 2010; Frumkin et al. Molecular Cell 2017). We study the connection between protein translation and protein folding where we show how programmed attenuation of ribosomes, in globular and membrane-embedded proteins, can facilitate folding (Yona et al eLife 2013; Fluman et al. eLife 2014). We study the economy of translation and the co-adaptation between the tRNA and mRNA pool in cells, mammals (Gingold et al. Cell 2014), and in microbes (Frumkin et al. PNAS 2018). In particular we discovered how distinct encoding programs of genes involved in cellular proliferation and differentiations in mammals have shaped and coordinated codon usage patterns and tRNA expression, genome wide (Gingold et al. Cell 2014). Such programs, we showed, operate also when cells undergo reverse differentiation, during the iPS paradigm (Zviran et al. with Jacob Hanna, Cell Stem Cell 2018). We now employ CRISPR techniques for gene deletion in human cells to knock out tRNA genes to examine their effects of cell proliferation and differentiation. We have been able to show that indeed distinct sets of differentially regulated tRNAs support cellular proliferation and differentiation. We study translation fidelity – the factors, embedded in genes, or distributed within the cellular milieu that determine patterns and levels or translation errors. We found that such “phenotypic mistakes” are regulated and we study if they have a role in evolution. For that we devised a mass spectrometry based methodology to find all translation errors made in a cell (Mordret et al. Molecular Cell 2019) that identifies mistakenly translated peptides within proteins. This experiment that allowed us to corroborate Kinetic Proofreading theories on trade-off between translation speed and accuracy. We study how translation accuracy is regulated in cancer and in aging
Genome and Systems Biology