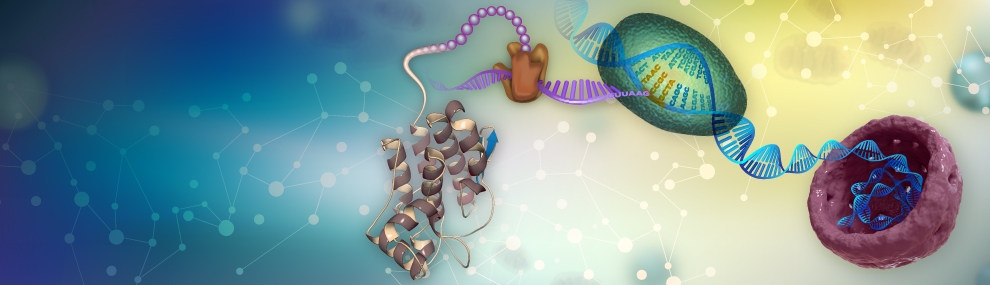
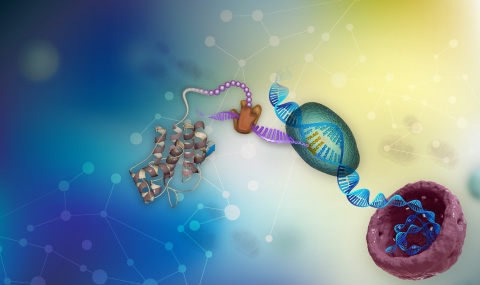
We use a diversity of research means and paradigms to study the mechanism and dynamics of genes’ and genome evolution. We evolve microbes in the lab towards several goals:
- We ask how organisms shape their genome as their environment changes (c.f. Yona et al. PNAS 2012), or when the genetics of their cellular machineries change, e.g. in translation (Yona et al. eLife 2013), or splicing (Frumkin et al. PLoS Biology 2019).
- We employ synthetic DNA libraries to decipher economy of gene expression (c.f. Frumkin et al. Molecular Cell 2017). We now expand the scope of study of economy of gene expression to study the evolutionary forces that shape cooperation between microbial cells, mainly through production and secretion of public good proteins.
- We develop sophisticated genetic means to study en mass mating selection and hybrid vigor among hundreds of yeast strains.
- We study evolution of evolvability (Yona et al. Cell 2015) by examining genetic and environmental means that confer upon organisms the capacity to evolve. Two particularly interesting evolvability mechanisms that we study are Horizontal Gene Transfer in bacteria, and Reverse Transcription in yeast. We organize Evolthon an international community challenge on lab evolution (Kaminski et al, PLoS Biology), a joint endeavor to investigate means for efficient lab evolution.
- We are interesting in the theory of evolution. Through mathematical models and computer simulations we study some basic aspects of evolutionary dynamics. Three prime questions that we ask along these lines are: what are optimal, potentially dynamic, regimes of change and inheritance of mutation rates among evolving and competing individuals? What is the role of phenotypic mutations in determining the trajectory of evolution? And, using mutation-selection-balance models we investigate alternative to Survival of The Fittest, e.g. cases in which the Quasi Species model predicts “Survival of the Flattest”.